Amino Acid Difference Formula to Help Explain Protein Evolution
Conversely a radical replacement or radical substitution is an amino acid replacement that exchanges an initial. Kyte J Doolittle RF.
Science 185 862-864 1974.
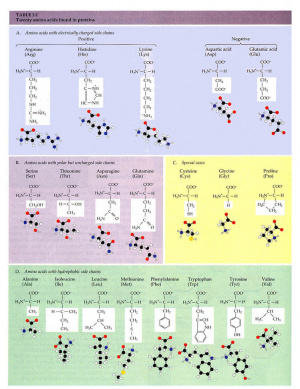
. Article for Grantham R. J Mol Biol 157 105-32 1982. The first protein to be sequenced was insulin which is composed of only 51 amino acids.
This formula was derived in 1974 from 3 amino acid physicochemical. Google Scholar Sneath PH. Science 185 862-864 1974.
Distance between amino acids a i and a j is calculated as where is value of polarity difference between replaced amino acids and and is difference for volume. J Biochem 88 1895-1898 1980. A formula for difference between amino acids combines properties that correlate best with protein residue substitution frequencies.
Thermostability and aliphatic index of globular proteins. The sequences analyzed included both prokaryotic and eukaryotic examples as well as mitochondrial cytochromeb and chloroplastb 6. 1974 Amino acid difference formula to help explain protein evolution.
The distance is based on three chemical properties of amino acid side chains. Has been cited by the following article. SGC table and Grantham difference formula.
A formula for difference between amino acids combines properties that correlate best with protein residue substitution frequencies. Scores amino acid polarity volume and lipophilicity. D i j ραc i c j 2 βp i p j 2 γv i v j 2 12.
Abstract for Amino acid difference formula to help explain protein evolution. Charge hydrophobicity and size. PMC free article Google Scholar Grantham R.
The amino acids such as hydrophobicity volume composition and secondary structure preferences. The alakazam package includes a set of functions to analyze the physicochemical properties of Ig and TCR amino acid sequences. 185 Amino Acid Difference Formula to Help Explain Protein Evolution Abstract.
J Biochem 88 1895-1898 1980. Proc Natl Acad Sci U S A. As an example for the SCIENCE VOL.
J Mol Biol 157 105-32 1982. Amino Acid Difference Formula to Help Explain Protein Evolution. A formula for diference between amino acids combines properties that correlate best with protein residue substitution frequencies.
Structural convergence during protein evolution. It has been demonstrated that the first two properties are the most important determinants in the folding of a protein and thus the dominant factors. Sanger and Thompson 1953.
Substitution frequencies agree much better with overall chemical difference between exchanging residues than with. This study explores the use of the Miyazawa-Jernigan residue contact potential in analyses of protein interaction and recognition between sense and complementary antisense peptides. Using position specific scoring matrix and auto covariance to predict protein subnuclear localization.
The same process can be applied to other regions simply by altering the sequence data column used. A substitution cysteine for tryptophan or of tryptophan for cysteine has a score of Any variation involving cysteine has a high or very high Grantham score and is predicted to be deleterious. Substitution frequencies agree much better with overall chemical difference between exchanging residues than with minimum base changes between their codons.
Role of the amino-acid code and of selection for conformation in the evolution of proteins. Substitution frequencies agree much better with overall chemical difference between exchanging residues than with minimum base changes between their codons. Thermostability and aliphatic index of globular proteins.
Composition polarity and molecular volume. Google Scholar Sneath PH. The classic results by Biro Blalock and Root-Bernstein link genetic code nucleotide patterns to amino acid properties protein structure and interaction.
Up to 10 cash back The amino acid sequences of the protonmotive cytochromeb from seven representative and phylogenetically diverse species have been compared to identify protein regions or segments that are conserved during evolution. A formula for difference between amino acids combines properties that correlate best with protein residue substitution frequencies. Substitution frequencies agree much better with overall chemical difference between.
Amino acid difference formula to help explain protein evolution. Amino acid difference formula to help explain protein evolution. Salemme FR Miller MD Jordan SR.
Of particular interest is the analysis of CDR3 properties which this vignette will demonstrate. Comparison of the amino acid sequence of insulin in a variety of mammals showed that with the exception of a stretch of three amino acids the protein is identical in each of the species. Kyte J Doolittle RF.
Amino Acid Difference Formula to Help Explain Protein Evolution. Composition polarity and molecular volume. A simple method for displaying the hydropathic character of a protein.
Amino acid difference formula to help explain protein evolution Science. Amino acid property analysis. A formula for difference between amino acids combines properties that correlate best with protein residue substitution frequencies.
The best second base amino acid score hierarchy U2 C2 G2 A2 shown in Table 4 was also compared to the amino acid properties of the Grantham difference formula Grantham 1974. Abstract for Amino acid difference formula to help explain protein evolution. Composition polarity and molecular volume.
A simple method for displaying the hydropathic character of a protein. A conservative replacement also called a conservative mutation or a conservative substitution is an amino acid replacement in a protein that changes a given amino acid to a different amino acid with similar biochemical properties eg. Substitution frequencies agree much better with.
Amino acid difference formula to help explain protein evolution. The Grantham distance d i j for two amino acids i and j is. Composition polarity and molecular volume.
And are standard deviations for and. Google Scholar Grantham R. Amino acid difference formula to help explain protein evolution.
Composition polarity and molecular volume. Examples of Protein Evolution. Composition c polarity p molecular volume v We provide a data set with these properties.
AMINO ACID DIFFERENCE FORMULA TO HELP EXPLAIN PROTEIN EVOLUTION.
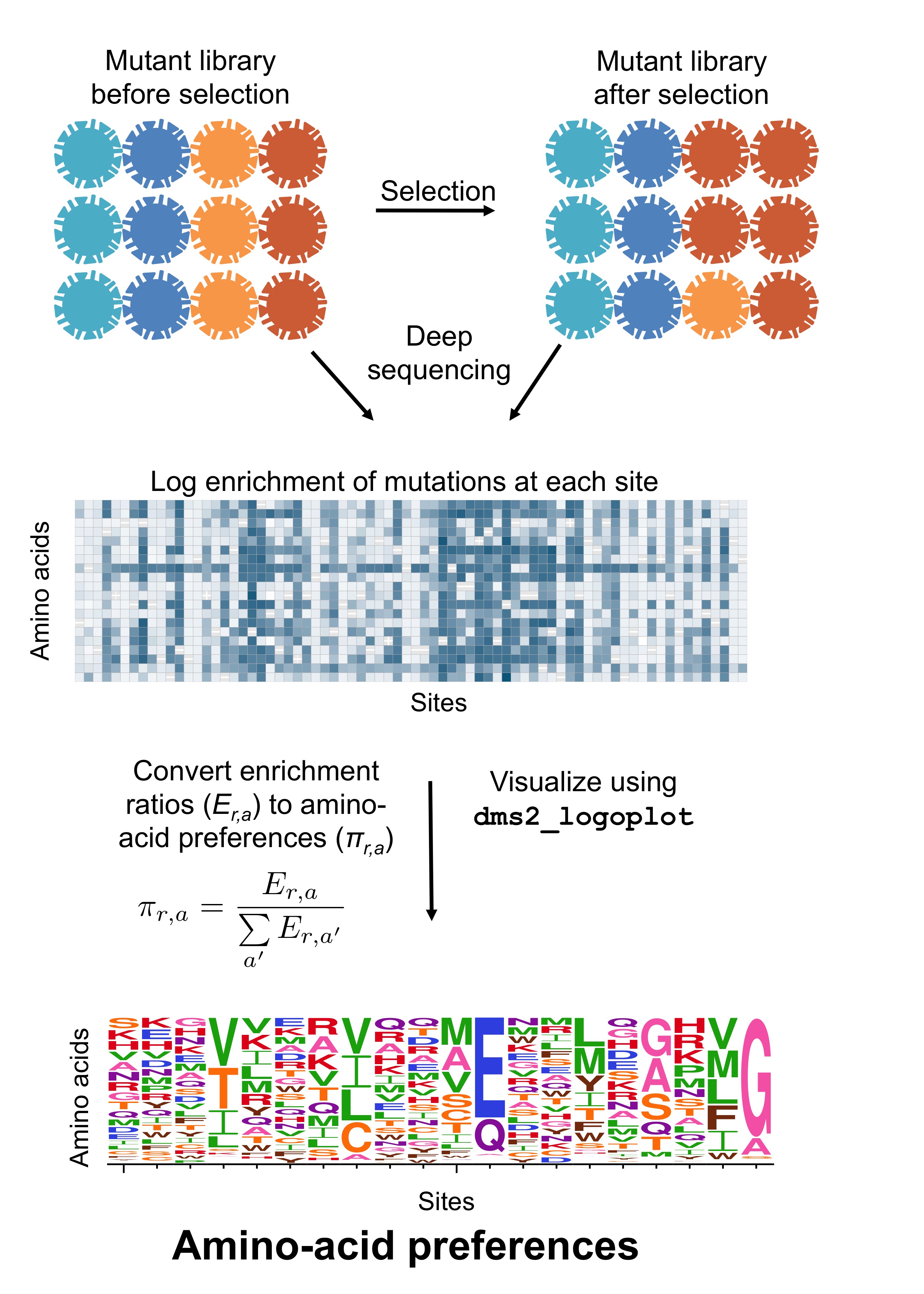
Amino Acid Preferences Dms Tools2 2 6 6 Documentation
Fischer Projection Formulae For Reference Glyceraldehydes A And Amino Download Scientific Diagram
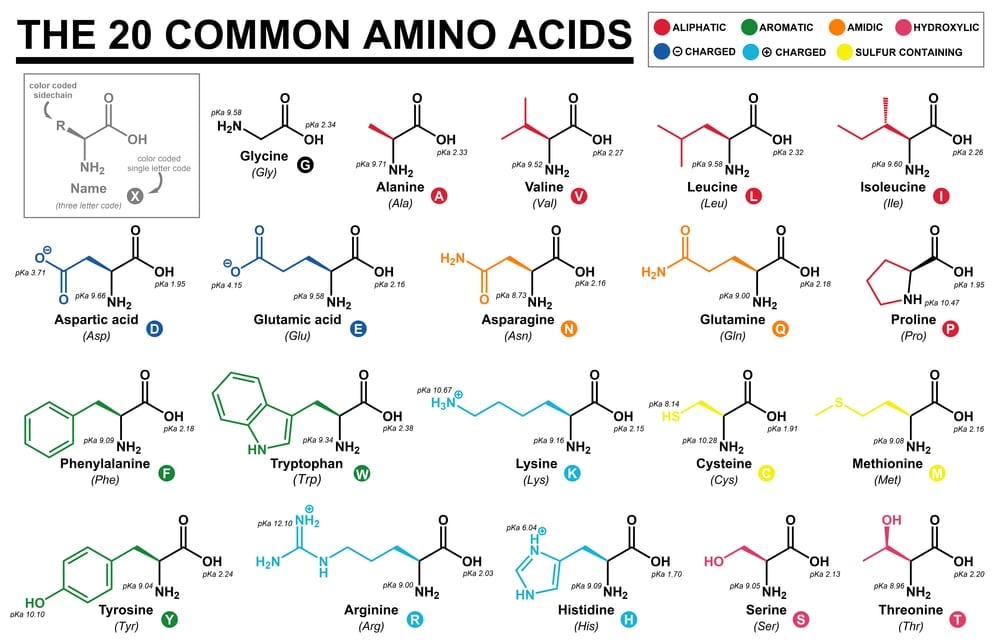
What Are The Two Rare Amino Acids Science Abc
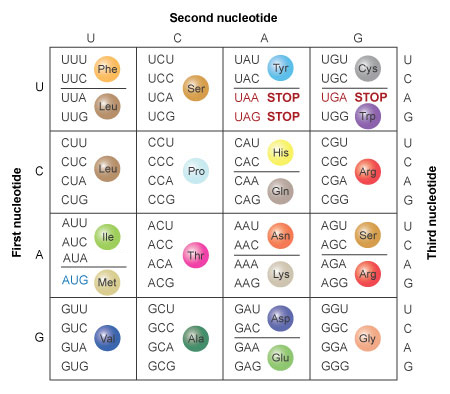
Nucleic Acids To Amino Acids Dna Specifies Protein Learn Science At Scitable
What Are The Main Differences Between Amino Acids And Proteins Quora
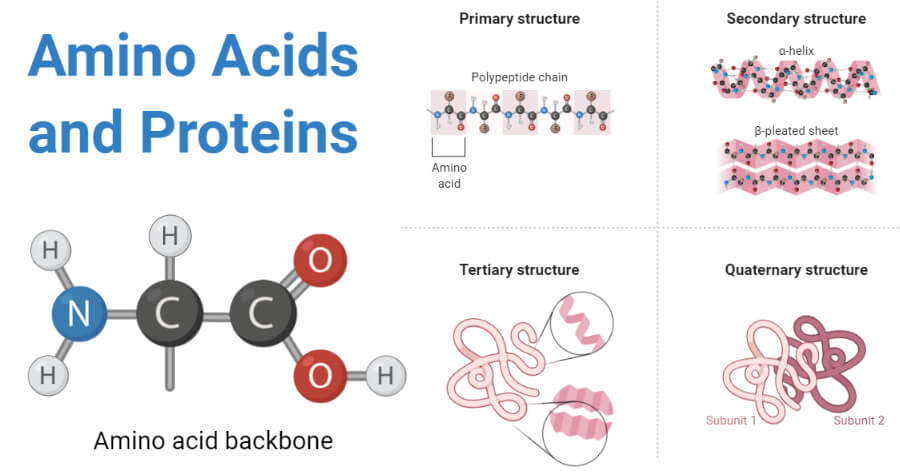
Amino Acids And Proteins Definition Structure Types Functions
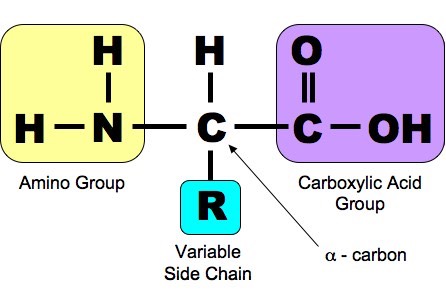


No comments for "Amino Acid Difference Formula to Help Explain Protein Evolution"
Post a Comment